2019年12月以来,中国湖北省武汉市部分医院陆续发现了多例不明原因肺炎病例,后被证实是由一种先前尚未发现的冠状病毒(coronavirus)感染引起的急性呼吸道传染病,这种病毒被世界卫生组织(World Health Organization, WHO)命名为2019新型冠状病毒(2019 novel coronavirus, 2019-nCoV)*[1]( *注:2020年2月11日,2019新型冠状病毒(2019-nCoV)被国际病毒分类委员会(the International Committee on Taxonomy of Viruses)冠状病毒研究小组(Coronavirus Study Group, CSG)命名为“SARS-CoV-2” (severe acute respiratory syndrome coronavirus 2),同时,由该病毒感染引起的疾病被WHO命名为“COVID-19” (corona virus disease 2019)。),该病毒与中东呼吸综合征相关冠状病毒(middle east respiratory syndrome-related coronavirus, MERSr-CoV)和严重急性呼吸综合征相关冠状病毒(severe acute respiratory syndrome-related coronavirus, SARSr-CoV)同属于β冠状病毒属[2]。
利用快速发展的基因组学方法与技术,全球的科研人员已经获得了多个2019-nCoV基因组序列,并且开展了多项相关研究[2,3,4,5,6,7]。因此,收集整合已有的2019-nCoV数据,构建统一完整的信息库系统,实现对数据的动态发布与共享对于防控病毒疫情、制定病毒性肺炎治疗方案具有重要意义[8,9]。自2020年1月5日,复旦大学张永振教授向美国国家生物技术信息中心(National Center for Biotechnology Information, NCBI)[10]的GenBank数据库提交第一条新型冠状病毒基因组序列(Acc. No. MN908947)至2020年2月5日,共有86条2019-nCoV序列数据在全球多个数据库发布,主要分布于德国全球流感病毒数据库(Global Initiative on Sharing All Influenza Data, GISAID)[11]、美国NCBI、深圳(国家)基因库(China National GeneBank, CNGB)[12]、国家微生物科学数据中心(National Microbiology Data Center, NMDC)[13]及国家生物信息中心(China National Center for Bioinformation, CNCB)/国家基因组科学数据中心(National Genomics Data Center, NGDC)[14]等相关数据库。然而,2019-nCoV序列数据分散在这些数据库中,未形成完整、统一访问的数据集,这给科研人员检索、预览和获取数据带来诸多不便。
为了缓解当前数据多源的局面和问题,帮助科研人员便捷地获取数据,同时提供高效的基因组序列递交与发布共享系统,CNCB/NGDC通过整合全球2019-nCoV相关数据,构建了2019新型冠状病毒信息库(2019nCoVR,
1 2019nCoVR数据资源
1.1 基因组序列发布动态
基于严格的质控审编流程,2019nCoVR收集整合多个数据平台(CNCB/NGDC、CNGB、GISAID、NCBI、NMDC)的2019-nCoV序列信息和元数据信息(包括病毒株名、序列号、数据来源、宿主、采样日期、采样地点、样本提供单位、数据递交单位等),持续更新序列发布动态,为开展相关科学研究提供完备准确的第一手数据。自2019年12月2019-nCoV疫情爆发至2020年2月5日,已收录来自16个国家/地区的81株病毒的86条基因组序列(附表1),其中67株具有全基因组序列(人体中分离66株,蝙蝠中分离1株)。
在数据获取与访问权限方面,遵守不同数据共享平台的数据管理规则,提供最大限度的数据集成与访问。可公开访问的数据已整合录入2019nCoVR,包括NCBI、CNCB/NGDC、NMDC、CNGB中相关基因组序列,任何人可不受限访问并下载;受限访问的数据,主要为GISAID数据库中序列,用户需到GISAID系统注册、登录后才可访问并下载(图1)。
图1
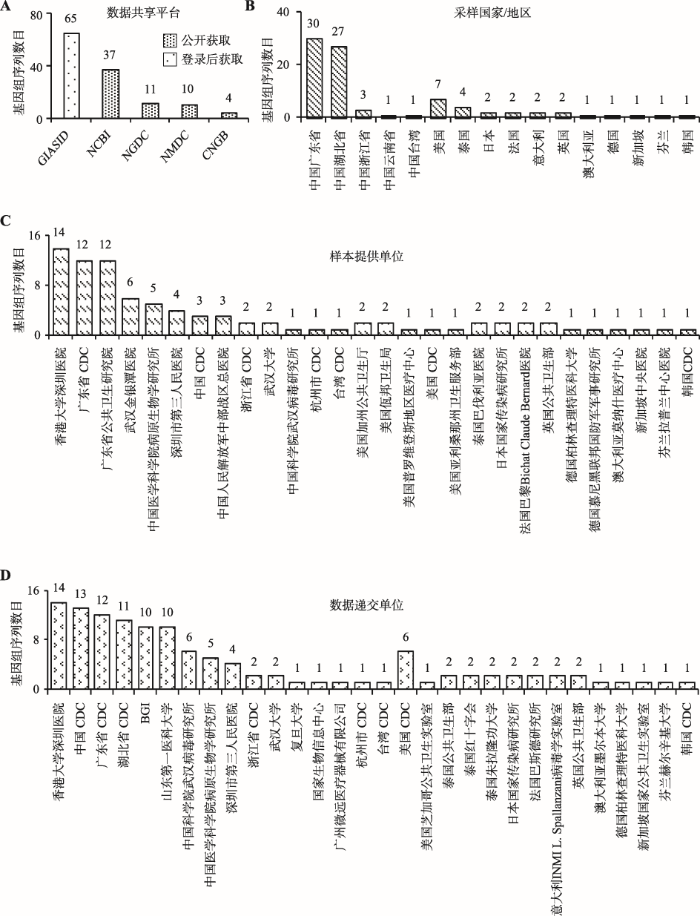
图1 2019新型冠状病毒基因组元信息相关统计结果
A:数据共享平台;B:采样国家/地区;C:样本提供单位;D:数据递交单位。
Fig. 1 Statistics of 2019-nCoV genome meta information
在病毒来源方面,所收录的病毒株主要来自湖北省武汉市,部分来自广东省和浙江省等地区,还有一小部分来自美国、泰国和日本等国家。病毒样本采集单位主要包括香港大学深圳医院、广东省疾病预防控制中心(Center for Disease Control and Prevention, CDC)、广东省公共卫生研究院、武汉金银潭医院、中国医学科学院病原生物学研究所等国内外28家医疗卫生或科研单位。基因组测序和数据递交主要由香港大学深圳医院、中国CDC、广东省CDC、湖北省CDC、华大基因(Beijing Genomics Institute, BGI)等30家单位完成(图1)。
1.2 基因组序列资源整合与信息检索
基于CNCB/NGDC的GWH数据平台,2019nCoVR收录并整合国内外公共数据平台中可开放获取的冠状病毒序列数据,形成冠状病毒序列数据集。截止到2020年2月5日,已审编收录冠状病毒科的核苷酸序列7566条和蛋白质序列29039条,以及相应的元数据信息(图2)。基于标准化的信息整合与发布,2019nCoVR提供多方位信息检索、条件查询、批量下载等功能,用户亦可在FTP网站公开访问和下载数据(
图2
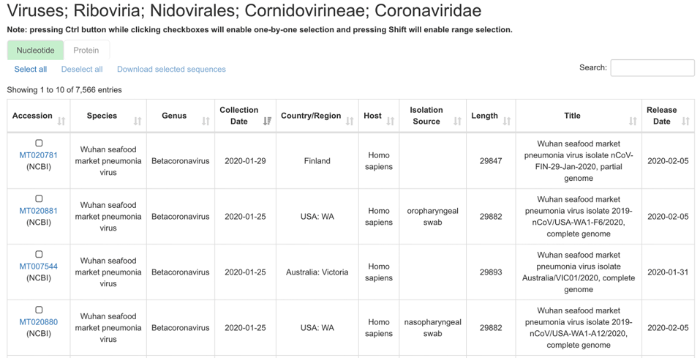
图2 冠状病毒科基因组序列信息汇总
Fig. 2 Coronaviridae genome sequence information
1.3 基因组序列变异分析与可视化
2019nCoVR分别选取可感染人的两种冠状病毒,即SARS (NC_004718)和最先公布的2019-nCoV基因组序列(MN908947),以及一种从蝙蝠中分离采集到的SARS样冠状病毒(bat-SL-CoVZC45, MG772933)作为参考基因组,整合“发布动态”中汇总可获取的全基因组序列,用Muscle软件[19,20]逐一进行全基因组序列比较和多序列比对,比较发现2019-nCoV与NC_004718、bat-SL-CoVZC45和蝙蝠中检测到的冠状病毒(bat/Yunnan/RaTG13/2013)的基因组序列相似度分别为80%、88%和96%,而2019-nCoV内部不同株系间的序列相似性约为99.9%。基于从人体中分离的65株病毒全基因组序列,在去除序列变异数量异常和变异位点集中(有5个突变发生在20 bp的区域内)的3株序列后,对剩余的62株序列采用基于距离的UPGMA法构建系统发育树,显示其遗传关系非常近且有所分化(图3)。
图3
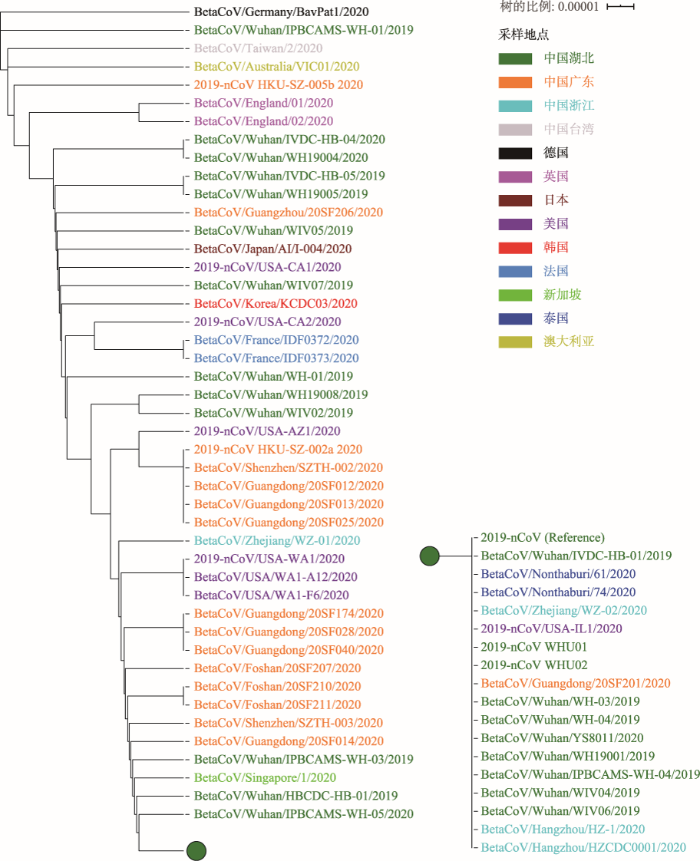
图3 新型冠状病毒序列的系统进化树
Fig. 3 Phylogenetics tree of 2019-nCoV
通过提取基因组序列比对中发现的变异位置、类型及信息,并配置GBrowse浏览器[21],可视化展示了每个病毒分离株与不同参考序列的变异(图4A)。此外,统计包括插入、删除、Indel和单核苷酸多态位点(SNP)的各类变异总数,提供了每个病毒株变异统计信息检索及下载。汇总各株变异信息发现主要的变异类型是SNP。经统计,与2019-nCoV参考序列相比,有14株病毒的序列无变异,49株平均有1~9个SNP变异(图4B),1株有27个SNP变异,因此推测该株(Acc. No. EPI_ISL_406592)的基因组序列质量存在问题。此外,检测到的少数序列删除变异(deletion)主要发生在基因组的5ʹUTR和3ʹUTR区域,有可能与测序准确率、基因组拼接等有关。初步提示已发布的65株病毒可能来源于近期出现的同一个病毒源。
图4
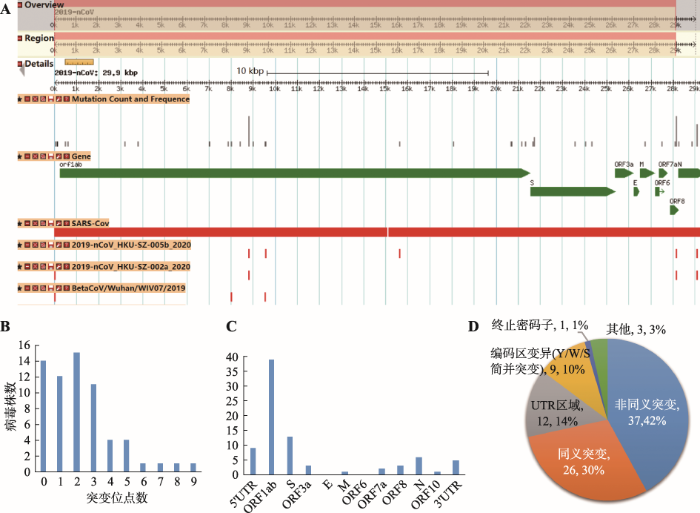
图4 基因组序列变异在线展示示意图及变异信息统计与注释
A:全基因组序列变异在线展示示意图;B:病毒株SNP变异数统计;C:SNP变异在各注释基因及UTR区的数量统计;D:SNP变异效应统计。
Fig. 4 Snapshot of genome sequence variants on GBrowse as well as SNP statistics and annotations
通过计算每个变异位点的群体发生频率并采用VEP软件[22]对上述变异进行注释,网站提供了所有变异位点注释信息(包括碱基变异、密码子及氨基酸变化、变异注释类型)的查询、浏览与下载。经统计发现2019-nCoV群体内的序列变异主要发生在5个基因,即产生病毒表面糖蛋白的S基因、编码病毒核衣壳磷蛋白的N基因、orf8基因、orf3a和最大的基因orf1ab。其中,orf1ab基因的变异位点数高达39 (图4C)。经分析,约42%的变异是非同义突变(图4D),且发现多株病毒的非同义突变主要发生在S蛋白的第32位(F→I,c.94Ttc>Atc)和第49位(H→Y,c.145Cat>Tat)及ORF8蛋白的第84位(L→S,c.251tTa>tCa),而发生在ORF1ab蛋白上的非同义突变位点数量最多(附表2)。
1.4 关联信息整合
2019nCoVR整合了来源于公共数据库及公共媒体的相关信息,主要包括:(1) NCBI冠状病毒科的所有序列、冠状病毒全基因组序列、感染人的冠状病毒全基因组序列、2019-nCoV序列等;(2)PubMed中冠状病毒相关的学术文献及Europe PMC针对2019-nCoV的最新学术报道;(3)中国CDC及WHO等权威机构对2019-nCoV的新闻报道、病毒解读及其相关的科普知识。这些内容为全球科研人员和普通民众开展学术研究、了解科研进展、掌握新闻动态与科学知识提供一站式数据资源与信息窗口。
2 数据汇交与审核机制
依托CNCB/NGDC的GSA系统,2019nCoVR提供新型冠状病毒原始测序数据的汇交服务,汇交内容主要包括元数据信息和序列文件。数据递交完成后,GSA系统会对用户递交的元数据信息和序列文件进行质量控制与审核,校验文件大小和内容、统计序列信息、评估数据质量,以此确保递交数据的完整性和可靠性。审核通过后,系统会为该数据分配唯一的数据编号(accession number),并通过邮件通知递交者。数据编号可作为数据检索和访问的标识,也可在文章中使用。
与之类似,2019nCoVR依托CNCB/NGDC的GWH数据库,汇交新型冠状病毒基因组序列和蛋白质序列,主要包括元数据、序列信息和注释文件。
为严格把控病毒基因组数据入库质量,针对用户递交的数据,GWH建立了严格的质量控制标准,审核检查数据的合法性和一致性,主要包括序列合法性、基因结构与信息完整性、基因结构内部的一致性、序列内容与注释信息的一致性以及载体、接头、index、污染序列等。数据审核通过后,GWH系统会为该数据分配正式的数据编号,方便数据检索、访问和下载。截止到2020年2月5日,已经收录了中国医学科学院病原生物学研究所和中国科学院武汉病毒研究所提交的11株冠状病毒全基因组序列。为了进一步扩大2019-nCoV基因组序列的国际影响力和应用范围,CNCB/NGDC与国际生物信息数据库建立了数据同步共享机制,第一批5个2019-nCoV全基因组序列已经在NCBI发布(Acc. No. MT019529~ MT019533)。
3 数据发布、管理与使用规范
2019nCoVR遵循CNCB/NGDC的相关数据管理制度及领域内数据管理惯例,即数据的所有权属于数据递交者,数据的公开与发布由数据提交者(submitter)自行管理。递交至2019nCoVR的新型冠状病毒数据(包括元信息和关联的序列数据),将分为公开(public)、受控(controlled)和私有(confidential)三种类型,数据提交者根据其数据的密级、保密期限、开放条件、开放对象和审核程序等,在提交数据时选择一种数据访问类型,数据提交完成并审核通过后,系统将按照数据提交者选择的数据访问类型进行管理(表1)。三类数据访问类型的管理规则具体如下:
(1)公开类型:元信息和关联序列数据都公开共享,任何用户可查询、访问与下载;
(2)受控类型:元信息公开共享,但关联序列数据受控访问。数据申请者须向数据提交者提出序列数据使用请求,由数据提交者向数据申请者发放访问权限。数据提交者可根据情况动态调整数据访问类型。
(3)私有类型:元信息和关联数据不会在数据平台上展示,用户无法查询、访问或下载。当私有数据符合开放共享条件时,如相关科研论文已发表或者达到约定公开时限,系统会通知数据提交者公开其数据。私有类型数据亦可由数据提交者动态管理。
表1 三类数据访问类型的基本规则
Table 1
| 数据类型 | 汇交内容 | 公开程度 | 开放对象 | 开放条件 |
|---|---|---|---|---|
| 公开* |
元信息 关联数据 |
公开 | 所有用户 | 审核通过即公开 |
| 受控 |
元信息 关联数据 |
公开 受控 |
所有用户 申请用户 |
相关科研论文已发表或达到约定公开时限 |
| 私有 |
元信息 关联数据 |
受控 | 无 | 相关科研论文已发表或达到约定公开时限 |
*:凡是类似2019-nCoV等涉及国家或全球公共卫生安全,呼吁基因序列数据在测序完成后第一时间采用“公开”数据类型开放共享。
4 结语与展望
2019nCoVR整合来自CNCB/NGDC、CNGB、GISAID、NCBI及NMDC 的新型冠状病毒数据资源,无缝对接CNCB/NGDC的相关数据库,为新型冠状病毒基因组数据的快速发布与开放共享提供公共平台,也为加速开展病毒分类溯源、基因组演化、快速检测、药物研发、新型肺炎的精准预防与治疗等研究提供重要基础。随着2019-nCoV科研工作的深入开展,2019nCoVR将持续更新并发布相关基因组序列及其元数据信息,为攻坚2019-nCoV提供数据保障与信息支撑。同时,特此呼吁科研人员和医务工作者加快推进2019-nCoV基因组数据的汇交、共享与发布,建立实现全球数据共同体,协同战胜病毒疫情。
致谢
该信息库由国家生物信息中心(CNCB)/国家基因组科学数据中心(NGDC)建设并维护。在建设过程中,得到了北京大学罗静初教授的支持和帮助,在此表示感谢!信息库所有数据来源于用户直接递交或国内外公共数据平台,包括GISAID、NCBI/GenBank、NMDC、CNGB/CNGBdb等(附表1),在此,对所有样本收集和数据递交的单位和个人表示感谢!
附录
附表1和附表2见网站电子版www.chinagene.cn。
附表1 病毒基因组元信息表。信息统计截至2020年2月5日。
Table S1
| Virus Isolate Name | Accession ID | Data Source | Related ID | Host | Sample Collection Date | Location | Nuc.Completeness | Originating Lab | Submitting Lab |
|---|---|---|---|---|---|---|---|---|---|
| BetaCoV/USA/WA1-F6/2020 | EPI_ISL_407215 | GISAID | Human | 2020-1-25 | USA / Washington | Complete | WA State Department of Health | Pathogen Discovery, Respiratory Viruses Branch, Division of Viral Diseases, Centers for Dieases Control and Prevention | |
| BetaCoV/Korea/KCDC03/2020 | EPI_ISL_407193 | GISAID | Human | 2020-1-25 | Korea / Gyeonggi-do | Complete | Korea Centers for Disease Control & Prevention (KCDC) Center for Laboratory Control of Infectious Diseases Division of Viral Diseases | Korea Centers for Disease Control & Prevention (KCDC) Center for Laboratory Control of Infectious Diseases Division of Viral Diseases | |
| BetaCoV/Japan/AI/I-004/2020 | EPI_ISL_407084 | GISAID | Human | 2020-1-25 | Japan / Aichi | Complete | Department of Virology III, National Institute of Infectious Diseases | Pathogen Genomics Center, National Institute of Infectious Diseases | |
| BetaCoV/USA/WA1-A12/2020 | EPI_ISL_407214 | GISAID | Human | 2020-1-25 | USA / Washington | Complete | WA State Department of Health | Pathogen Discovery, Respiratory Viruses Branch, Division of Viral Diseases, Centers for Dieases Control and Prevention | |
| BetaCoV/Singapore/1/2020 | EPI_ISL_406973 | GISAID | Human | 2020-1-23 | Singapore | Complete | Singapore General Hospital | National Public Health Laboratory | |
| BetaCoV/England/02/2020 | EPI_ISL_407073 | GISAID | Human | 2020-1-29 | England | Complete | Respiratory Virus Unit, Microbiology Services Colindale, Public Health England | Respiratory Virus Unit, Microbiology Services Colindale, Public Health England | |
| BetaCoV/England/01/2020 | EPI_ISL_407071 | GISAID | Human | 2020-1-29 | England | Complete | Respiratory Virus Unit, Microbiology Services Colindale, Public Health England | Respiratory Virus Unit, Microbiology Services Colindale, Public Health England | |
| BetaCoV/Finland/1/2020 | EPI_ISL_407079 | GISAID | Human | 2020-1-29 | Finland / Lapland | Partial/scaffold level | Lapland Central Hospital | Department of Virology, University of Helsinki and Helsinki University Hospital, Helsinki, Finland | |
| BetaCoV/Hangzhou/HZ-1/2020 | EPI_ISL_406970 | GISAID | Human | 2020-1-20 | China / Zhejiang / Hangzhou | Complete | Hangzhou Center for Disease Control and Prevention, Microbiology Lab | Hangzhou Center for Disease Control and Prevention, Microbiology Lab | |
| 2019 nCoV/Italy-INMI1 | MT008022 | GenBank | EPI_ISL_406959 | Human | 2020-01 | Italy / Rome | Partial/gene level | Virology Laboratory, INMI L. Spallanzani | |
| 2019 nCoV/Italy-INMI2 | MT008023 | GenBank | EPI_ISL_406960 | Human | 2020-01 | Italy / Rome | Partial/gene level | Virology Laboratory, INMI L. Spallanzani | |
| BetaCoV/Wuhan/WH19002/2019 | NMDC60013002-05 | NMDC | Human | 2019-12-30 | China / Hubei Province / Wuhan City | Complete | China CDC; Shandong First Medical University & Shandong Academy of Medical Sciences; Hubei Provincial Center for Disease Control and Prevention; BGI PathoGenesis Pharmaceutical Technology Co., Ltd | ||
| BetaCoV/Wuhan/WH19008/2019 | NMDC60013002-06 | NMDC | Human | 2019-12-30 | China / Hubei Province / Wuhan City | Complete | China CDC; Shandong First Medical University & Shandong Academy of Medical Sciences; Hubei Provincial Center for Disease Control and Prevention; BGI PathoGenesis Pharmaceutical Technology Co., Ltd | ||
| BetaCoV/Wuhan/YS8011/2020 | NMDC60013002-07 | NMDC | Human | 2020-1-7 | China / Hubei Province / Wuhan City | Complete | China CDC; Shandong First Medical University & Shandong Academy of Medical Sciences; Hubei Provincial Center for Disease Control and Prevention; BGI PathoGenesis Pharmaceutical Technology Co., Ltd | ||
| BetaCoV/Wuhan/WH19001/2019 | NMDC60013002-08 | NMDC | Human | 2019-12-30 | China / Hubei Province / Wuhan City | Complete | China CDC; Shandong First Medical University & Shandong Academy of Medical Sciences; Hubei Provincial Center for Disease Control and Prevention; BGI PathoGenesis Pharmaceutical Technology Co., Ltd | ||
| BetaCoV/Wuhan/WH19004/2020 | NMDC60013002-09 | NMDC | Human | 2020-1-1 | China / Hubei Province / Wuhan City | Complete | China CDC; Shandong First Medical University & Shandong Academy of Medical Sciences; Hubei Provincial Center for Disease Control and Prevention; BGI PathoGenesis Pharmaceutical Technology Co., Ltd | ||
| BetaCoV/Wuhan/WH19005/2019 | NMDC60013002-10 | NMDC | Human | 2019-12-30 | China / Hubei Province / Wuhan City | Complete | China CDC; Shandong First Medical University & Shandong Academy of Medical Sciences; Hubei Provincial Center for Disease Control and Prevention; BGI PathoGenesis Pharmaceutical Technology Co., Ltd | ||
| BetaCoV/Germany/BavPat1/2020 | EPI_ISL_406862 | GISAID | Human | 2020-1-28 | Germany / Bavaria / Munich | Complete | Charité Universitätsmedizin Berlin, Institute of Virology; Institut für Mikrobiologie der Bundeswehr, Munich | Charité Universitätsmedizin Berlin, Institute of Virology | |
| BetaCoV/Wuhan/WH-01/2019 | NMDC60013002-01 | NMDC | LR757998, EPI_ISL_406798, CNA0007332 | Human | 2019-12-26 | China / Hubei Province / Wuhan City | Complete | General Hospital of Central Theater Command of People's Liberation Army of China | BGI PathoGenesis Pharmaceutical Technology Co., Ltd; China CDC; Shandong First Medical University & Shandong Academy of Medical Sciences; Hubei Provincial Center for Disease Control and Prevention |
| BetaCoV/Wuhan/WH-02/2019 | NMDC60013002-02 | NMDC | LR757997, CNA0007333 | Human | 2019-12-31 | China / Hubei Province / Wuhan City | Partial/scaffold level | BGI PathoGenesis Pharmaceutical Technology Co., Ltd; China CDC; Shandong First Medical University & Shandong Academy of Medical Sciences; Hubei Provincial Center for Disease Control and Prevention | |
| BetaCoV/Wuhan/WH-03/2019 | NMDC60013002-03 | NMDC | LR757996, EPI_ISL_406800, CNA0007334 | Human | 2020-1-1 | China / Hubei Province / Wuhan City | Complete | General Hospital of Central Theater Command of People's Liberation Army of China | BGI PathoGenesis Pharmaceutical Technology Co., Ltd; China CDC; Shandong First Medical University & Shandong Academy of Medical Sciences; Hubei Provincial Center for Disease Control and Prevention |
| BetaCoV/Wuhan/WH-04/2019 | NMDC60013002-04 | NMDC | LR757995, EPI_ISL_406801, CNA0007335 | Human | 2020-1-5 | China / Hubei Province / Wuhan City | Complete | General Hospital of Central Theater Command of People's Liberation Army of China | BGI PathoGenesis Pharmaceutical Technology Co., Ltd; China CDC; Shandong First Medical University & Shandong Academy of Medical Sciences; Hubei Provincial Center for Disease Control and Prevention |
| BetaCoV/Australia/VIC01/2020 | MT007544 | GenBank | EPI_ISL_406844 | Human | 2020-1-25 | Australia / Victoria / Clayton | Complete | Monash Medical Centre | Department of Microbiology and Immunology, The University of Melbourne at The Peter Doherty Institute for Infection and Immunity |
| WIV02 | GWHABKK00000000 | Genome Warehouse | EPI_ISL_402127, MN996527 | Human | 2019-12-30 | China/ Hubei / Wuhan | Complete | Wuhan Jinyintan Hospital | CAS Key Laboratory of Special Pathogens and Biosafety and Center for Emerging Infectious Diseases, Wuhan Institute of Virology, Chinese Academy of Sciences |
| WIV04 | GWHABKL00000000 | Genome Warehouse | EPI_ISL_402124, MN996528 | Human | 2019-12-30 | China / Hubei / Wuhan | Complete | Wuhan Jinyintan Hospital | CAS Key Laboratory of Special Pathogens and Biosafety and Center for Emerging Infectious Diseases, Wuhan Institute of Virology, Chinese Academy of Sciences |
| WIV05 | GWHABKM00000000 | Genome Warehouse | EPI_ISL_402128, MN996529 | Human | 2019-12-30 | China / Hubei / Wuhan | Complete | Wuhan Jinyintan Hospital | CAS Key Laboratory of Special Pathogens and Biosafety and Center for Emerging Infectious Diseases, Wuhan Institute of Virology, Chinese Academy of Sciences |
| WIV06 | GWHABKN00000000 | Genome Warehouse | EPI_ISL_402129, MN996530 | Human | 2019-12-30 | China / Hubei / Wuhan | Complete | Wuhan Jinyintan Hospital | CAS Key Laboratory of Special Pathogens and Biosafety and Center for Emerging Infectious Diseases, Wuhan Institute of Virology, Chinese Academy of Sciences |
| WIV07 | GWHABKO00000000 | Genome Warehouse | EPI_ISL_402130, MN996531 | Human | 2019-12-30 | China / Hubei / Wuhan | Complete | Wuhan Jinyintan Hospital | CAS Key Laboratory of Special Pathogens and Biosafety and Center for Emerging Infectious Diseases, Wuhan Institute of Virology, Chinese Academy of Sciences |
| TG13 | GWHABKP00000000 | Genome Warehouse | EPI_ISL_402131, MN996532 | Rhinolophus affinis | 2013-7-24 | China / Yunnan / Pu'er | Complete | Wuhan Institute of Virology | CAS Key Laboratory of Special Pathogens and Biosafety and Center for Emerging Infectious Diseases, Wuhan Institute of Virology, Chinese Academy of Sciences |
| BetaCoV/Guangdong/20SF174/2020 | EPI_ISL_406531 | GISAID | Human | 2020-1-22 | China / Guangdong Province | Complete | Guangdong Provincial Center for Diseases Control and Prevention; Guangdong Provincial Institute of Public Health | Guangdong Provincial Center for Diseases Control and Prevention | |
| BetaCoV/Guangzhou/20SF206/2020 | EPI_ISL_406533 | GISAID | Human | 2020-1-22 | China / Guangdong Province / Guangzhou City | Complete | Guangdong Provincial Center for Diseases Control and Prevention; Guangdong Provincial Institute of Public Health | Guangdong Provincial Center for Diseases Control and Prevention | |
| BetaCoV/Foshan/20SF207/2020 | EPI_ISL_406534 | GISAID | Human | 2020-1-22 | China / Guangdong Province | Complete | Guangdong Provincial Center for Diseases Control and Prevention; Guangdong Provincial Institute of Public Health | Guangdong Provincial Center for Diseases Control and Prevention | |
| BetaCoV/Foshan/20SF210/2020 | EPI_ISL_406535 | GISAID | Human | 2020-1-22 | China / Guangdong Province | Complete | Guangdong Provincial Center for Diseases Control and Prevention; Guangdong Provincial Institute of Public Health | Guangdong Provincial Center for Diseases Control and Prevention | |
| BetaCoV/Foshan/20SF211/2020 | EPI_ISL_406536 | GISAID | Human | 2020-1-22 | China / Guangdong Province | Complete | Guangdong Provincial Center for Diseases Control and Prevention; Guangdong Provincial Institute of Public Health | Guangdong Provincial Center for Diseases Control and Prevention | |
| BetaCoV/Guangdong/20SF201/2020 | EPI_ISL_406538 | GISAID | Human | 2020-1-23 | China / Guangdong Province | Complete | Guangdong Provincial Center for Diseases Control and Prevention;Guangdong Provincial Institute of Public Health | Guangdong Provincial Center for Diseases Control and Prevention | |
| BetaCoV/Shenzhen/SZTH-001/2020 | EPI_ISL_406592 | GISAID | Human | 2020-1-13 | China/ Guangdong Province / Shenzhen City | Complete | Shenzhen Third People's Hospital | Shenzhen Key Laboratory of Pathogen and Immunity, National Clinical Research Center for Infectious Disease, Shenzhen Third People's Hospital | |
| BetaCoV/Shenzhen/SZTH-003/2020 | EPI_ISL_406594 | GISAID | Human | 2020-1-16 | China/ Guangdong Province / Shenzhen City | Complete | Shenzhen Key Laboratory of Pathogen and Immunity, National Clinical Research Center for Infectious Disease, Shenzhen Third People's Hospital | Shenzhen Key Laboratory of Pathogen and Immunity, National Clinical Research Center for Infectious Disease, Shenzhen Third People's Hospital | |
| BetaCoV/Shenzhen/SZTH-004/2020 | EPI_ISL_406595 | GISAID | Human | 2020-1-16 | China/ Guangdong Province / Shenzhen City | Complete | Shenzhen Key Laboratory of Pathogen and Immunity, National Clinical Research Center for Infectious Disease, Shenzhen Third People's Hospital | Shenzhen Key Laboratory of Pathogen and Immunity, National Clinical Research Center for Infectious Disease, Shenzhen Third People's Hospital | |
| BetaCoV/Shenzhen/SZTH-002/2020 | EPI_ISL_406593 | GISAID | Human | 2020-1-13 | China/ Guangdong Province / Shenzhen City | Complete | Shenzhen Key Laboratory of Pathogen and Immunity, National Clinical Research Center for Infectious Disease, Shenzhen Third People's Hospital | Shenzhen Key Laboratory of Pathogen and Immunity, National Clinical Research Center for Infectious Disease, Shenzhen Third People's Hospital | |
| BetaCoV/France/IDF0373/2020 | EPI_ISL_406597 | GISAID | Human | 2020-1-23 | France / Ile-de-France / Paris | Complete | Department of Infectious and Tropical Diseases, Bichat Claude Bernard Hospital, Paris | National Reference Center for Viruses of Respiratory Infections, Institut Pasteur, Paris | |
| BetaCoV/France/IDF0372/2020 | EPI_ISL_406596 | GISAID | Human | 2020-1-23 | France / Ile-de-France / Paris | Complete | Department of Infectious and Tropical Diseases, Bichat Claude Bernard Hospital, Paris | National Reference Center for Viruses of Respiratory Infections, Institut Pasteur, Paris | |
| 2019-nCoV_HKU-SZ-001_2020 | MN938387 | GenBank | Human | 2020-01 | China / Guangdong / Shenzhen | Partial/gene level | University of Hong Kong-Shenzhen Hospital | University of Hong Kong-Shenzhen Hospital | |
| 2019-nCoV_HKU-SZ-001_2020 | MN938385 | GenBank | Human | 2020-01 | China / Guangdong / Shenzhen | Partial/gene level | University of Hong Kong-Shenzhen Hospital | University of Hong Kong-Shenzhen Hospital | |
| 2019-nCoV_HKU-SZ-002b_2020 | MN938388 | GenBank | Human | 2020-01 | China / Guangdong / Shenzhen | Partial/gene level | University of Hong Kong-Shenzhen Hospital | University of Hong Kong-Shenzhen Hospital | |
| 2019-nCoV_HKU-SZ-004_2020 | MN938389 | GenBank | Human | 2020-01 | China / Guangdong / Shenzhen | Partial/gene level | University of Hong Kong-Shenzhen Hospital | University of Hong Kong-Shenzhen Hospital | |
| 2019-nCoV_HKU-SZ-004_2020 | MN938386 | GenBank | Human | 2020-01 | China / Guangdong / Shenzhen | Partial/gene level | University of Hong Kong-Shenzhen Hospital | University of Hong Kong-Shenzhen Hospital | |
| 2019-nCoV_HKU-SZ-005_2020 | MN938390 | GenBank | Human | 2020-01 | China / Guangdong / Shenzhen | Partial/gene level | University of Hong Kong-Shenzhen Hospital | University of Hong Kong-Shenzhen Hospital | |
| 2019-nCoV_HKU-SZ-007a_2020 | MN975266 | GenBank | Human | 2020-01 | China / Guangdong / Shenzhen | Partial/gene level | University of Hong Kong-Shenzhen Hospital | University of Hong Kong-Shenzhen Hospital | |
| 2019-nCoV_HKU-SZ-007a_2020 | MN975263 | GenBank | Human | 2020-01 | China / Guangdong / Shenzhen | Partial/gene level | University of Hong Kong-Shenzhen Hospital | University of Hong Kong-Shenzhen Hospital | |
| 2019-nCoV_HKU-SZ-007b_2020 | MN975264 | GenBank | Human | 2020-01 | China / Guangdong / Shenzhen | Partial/gene level | University of Hong Kong-Shenzhen Hospital | University of Hong Kong-Shenzhen Hospital | |
| 2019-nCoV_HKU-SZ-007b_2020 | MN975267 | GenBank | Human | 2020-01 | China / Guangdong / Shenzhen | Partial/gene level | University of Hong Kong-Shenzhen Hospital | University of Hong Kong-Shenzhen Hospital | |
| 2019-nCoV_HKU-SZ-007c_2020 | MN975268 | GenBank | Human | 2020-01 | China / Guangdong / Shenzhen | Partial/gene level | University of Hong Kong-Shenzhen Hospital | University of Hong Kong-Shenzhen Hospital | |
| 2019-nCoV_HKU-SZ-007c_2020 | MN975265 | GenBank | Human | 2020-01 | China / Guangdong / Shenzhen | Partial/gene level | University of Hong Kong-Shenzhen Hospital | University of Hong Kong-Shenzhen Hospital | |
| SI200040-SP | MN970003 | GenBank | Human | 2020-1-8 | Thailand | Partial/gene level | Faculty of Medicine, Chulalongkorn University | ||
| SI200121-SP | MN970004 | GenBank | Human | 2020-1-13 | Thailand | Partial/gene level | Faculty of Medicine, Chulalongkorn University | ||
| BetaCoV/Taiwan/2/2020 | EPI_ISL_406031 | GISAID | Human | 2020-1-23 | Taiwan/Kaohsiung City | Complete | Centers for Disease Control (Taiwan) | Centers for Disease Control (Taiwan) | |
| 2019-nCoV/USA-CA1/2020 | MN994467 | GenBank | EPI_ISL_406034 | Human | 2020-1-23 | USA / California / Los Angeles | Complete | California Department of Public Health | Division of Viral Diseases, Centers for Disease Control and Prevention |
| 2019-nCoV/USA-CA2/2020 | MN994468 | GenBank | EPI_ISL_406036 | Human | 2020-1-22 | USA / California / Orange County | Complete | California Department of Public Health | Division of Viral Diseases, Centers for Disease Control and Prevention |
| 2019-nCoV/USA-AZ1/2020 | MN997409 | GenBank | EPI_ISL_406223 | Human | 2020-1-22 | USA / Arizona / Phoenix | Complete | Arizona Department of Health Services | Division of Viral Diseases, Centers for Disease Control and Prevention |
| 2019-nCoV WHU01 | MN988668 | GenBank | EPI_ISL_406716 | Human | 2020-1-2 | China / Hubei / Wuhan | Complete | State Key Laboratory of Virology, Wuhan University | State Key Laboratory of Virology, Wuhan University |
| 2019-nCoV WHU02 | MN988669 | GenBank | EPI_ISL_406717 | Human | 2020-1-2 | China / Hubei / Wuhan | Complete | State Key Laboratory of Virology, Wuhan University | State Key Laboratory of Virology, Wuhan University |
| 2019-nCoV/USA-WA1/2020 | MN985325 | GenBank | EPI_ISL_404895 | Human | 2020-1-19 | USA / Washington / Snohomish County | Complete | Providence Regional Medical Center | Division of Viral Diseases, Centers for Disease Control and Prevention |
| 2019-nCoV/USA-IL1/2020 | MN988713 | GenBank | EPI_ISL_404253 | Human | 2020-1-21 | USA / Illinois /Chicago | Complete | Pathogen Discovery, Respiratory Viruses Branch, Division of Viral Diseases, Centers for Dieases Control and Prevention | IL Department of Public Health Chicago Laboratory |
| BetaCoV/Wuhan/IPBCAMS-WH-01/2019 | GWHABKF00000000 | Genome Warehouse | EPI_ISL_402123 | Human | 2019-12-23 | China / Hubei Province / Wuhan City | Complete | Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College | Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College; Vision Medicals Co., Ltd |
| BetaCoV/Wuhan/IPBCAMS-WH-02/2019 | GWHABKG00000000 | Genome Warehouse | EPI_ISL_403931 | Human | 2019-12-30 | China / Hubei Province / Wuhan City | Complete | Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College | Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College |
| BetaCoV/Wuhan/IPBCAMS-WH-03/2019 | GWHABKH00000000 | Genome Warehouse | EPI_ISL_403930 | Human | 2019-12-30 | China / Hubei Province / Wuhan City | Complete | Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College | Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College |
| BetaCoV/Wuhan/IPBCAMS-WH-04/2019 | GWHABKI00000000 | Genome Warehouse | EPI_ISL_403929 | Human | 2019-12-30 | China / Hubei Province / Wuhan City | Complete | Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College | Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College |
| BetaCoV/Wuhan/IPBCAMS-WH-05/2020 | GWHABKJ00000000 | Genome Warehouse | EPI_ISL_403928 | Human | 2020-1-1 | China / Hubei Province / Wuhan City | Complete | Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College | Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College; China National Center for Bioinformation |
| 2019-nCoV_HKU-SZ-002a_2020 | MN938384 | GenBank | EPI_ISL_406030 | Human | 2020-1 | China / Guangdong / Shenzhen | Complete | University of Hong Kong-Shenzhen Hospital | University of Hong Kong-Shenzhen Hospital |
| 2019-nCoV_HKU-SZ-005b_2020 | MN975262 | GenBank | EPI_ISL_405839 | Human | 2020-1 | China / Guangdong / Shenzhen | Complete | University of Hong Kong-Shenzhen Hospital | University of Hong Kong-Shenzhen Hospital |
| BetaCoV/Guangdong/20SF040/2020 | EPI_ISL_403937 | GISAID | Human | 2020-1-18 | China / Guangdong Province / Zhuhai City | Complete | Guangdong Provincial Center for Diseases Control and Prevention; Guangdong Provincial Institute of Public Health | Department of Microbiology, Guangdong Provincial Center for Diseases control and Prevention | |
| BetaCoV/Guangdong/20SF028/2020 | EPI_ISL_403936 | GISAID | Human | 2020-1-17 | China / Guangdong Province / Zhuhai City | Complete | Guangdong Provincial Center for Diseases control and Prevention; Guangdong Provincial Institute of Public Health | Department of Microbiology, Guangdong Provincial Center for Diseases control and Prevention | |
| BetaCoV/Guangdong/20SF025/2020 | EPI_ISL_403935 | GISAID | Human | 2020-1-15 | China / Guangdong Province / Shenzhen City | Complete | Guangdong Provincial Center for Diseases control and Prevention; Guangdong Provincial Institute of Public Health | Department of Microbiology, Guangdong Provincial Center for Diseases control and Prevention | |
| BetaCoV/Guangdong/20SF014/2020 | EPI_ISL_403934 | GISAID | Human | 2020-1-15 | China / Guangdong Province / Shenzhen City | Complete | Guangdong Provincial Center for Diseases control and Prevention; Guangdong Provincial Institute of Public Health | Department of Microbiology, Guangdong Provincial Center for Diseases control and Prevention | |
| BetaCoV/Guangdong/20SF013/2020 | EPI_ISL_403933 | GISAID | Human | 2020-1-15 | China / Guangdong Province / Shenzhen City | Complete | Guangdong Provincial Center for Diseases control and Prevention; Guangdong Provincial Institute of Public Health | Department of Microbiology, Guangdong Provincial Center for Diseases control and Prevention | |
| BetaCoV/Guangdong/20SF012/2020 | EPI_ISL_403932 | GISAID | Human | 2020-1-14 | China / Guangdong Province / Shenzhen City | Complete | Guangdong Provincial Center for Diseases control and Prevention; Guangdong Provincial Institute of Public Health | Department of Microbiology, Guangdong Provincial Center for Diseases control and Prevention | |
| BetaCoV/Zhejiang/WZ-01/2020 | EPI_ISL_404227 | GISAID | Human | 2020-1-16 | China / Zhejiang Province | Complete | Zhejiang Provincial Center for Disease Control and Prevention | Department of Microbiology, Zhejiang Provincial Center for Disease Control and Prevention | |
| BetaCoV/Zhejiang/WZ-02/2020 | EPI_ISL_404228 | GISAID | Human | 2020-1-17 | China / Zhejiang Province | Complete | Zhejiang Provincial Center for Disease Control and Prevention | Department of Microbiology, Zhejiang Provincial Center for Disease Control and Prevention | |
| BetaCoV/Wuhan/HBCDC-HB-01/2019 | EPI_ISL_402132 | GISAID | Human | 2019-12-30 | China/Hubei Province | Complete | Wuhan Jinyintan Hospital | Hubei Provincial Center for Disease Control and Prevention | |
| BetaCoV/Nonthaburi/74/2020 | EPI_ISL_403963 | GISAID | Human | 2020-1-13 | Thailand/ Nonthaburi Province | Complete | Bamrasnaradura Hospital | Department of Medical Sciences, Ministry of Public Health, Thailand; Thai Red Cross Emerging Infectious Diseases - Health Science Centre; Department of Disease Control, Ministry of Public Health, Thailand | |
| BetaCoV/Nonthaburi/61/2020 | EPI_ISL_403962 | GISAID | Human | 2020-1-8 | Thailand/ Nonthaburi Province | Complete | Bamrasnaradura Hospital | Department of Medical Sciences, Ministry of Public Health, Thailand; Thai Red Cross Emerging Infectious Diseases - Health Science Centre; Department of Disease Control, Ministry of Public Health, Thailand | |
| BetaCoV/Wuhan/IVDC-HB-04/2020 | EPI_ISL_402120 | GISAID | Human | 2020-1-1 | China / Hubei Province / Wuhan City | Complete | National Institute for Viral Disease Control and Prevention, China CDC | National Institute for Viral Disease Control and Prevention, China CDC | |
| BetaCoV/Wuhan/IVDC-HB-01/2019 | EPI_ISL_402119 | GISAID | Human | 2019-12-30 | China / Hubei Province / Wuhan City | Complete | National Institute for Viral Disease Control and Prevention, China CDC | National Institute for Viral Disease Control and Prevention, China CDC | |
| BetaCoV/Wuhan/IVDC-HB-05/2019 | EPI_ISL_402121 | GISAID | Human | 2019-12-30 | China / Hubei Province / Wuhan City | Complete | National Institute for Viral Disease Control and Prevention, China CDC | National Institute for Viral Disease Control and Prevention, China CDC | |
| BetaCoV/Kanagawa/1/2020 | EPI_ISL_402126 | GISAID | Human | 2020-1-14 | Kanagawa Prefecture / Japan | Partial | Dept. of Virology III, National Institute of Infectious Diseases | Dept. of Virology III, National Institute of Infectious Diseases | |
| Wuhan-Hu-1 | MN908947 | GenBank | NC_045512 | Human | 2019-12 | China / Hubei Province / Wuhan City | Complete | Shanghai Public Health Clinical Center & School of Public Health, Fudan University, Shanghai, China | |
附表2 2019-nCoV基因组序列变异注释信息
Table S2
| 基因组位置 | 基因/区域名称 | 变异病毒株数 | 碱基变化及病毒株数 | 变异注释类型 | 蛋白名称.位置.氨基酸变化 | 基因名.CDS位置.序列变化 | 效应类型 |
|---|---|---|---|---|---|---|---|
| 2019-nCoV_16 | 5'UTR | 1 | C->T:1 | intergenic_variant | - | - | MODIFIER |
| 2019-nCoV_31 | 5'UTR | 1 | A->G:1 | intergenic_variant | - | - | MODIFIER |
| 2019-nCoV_104 | 5'UTR | 1 | T->A:1 | intergenic_variant | - | - | MODIFIER |
| 2019-nCoV_111 | 5'UTR | 1 | T->C:1 | intergenic_variant | - | - | MODIFIER |
| 2019-nCoV_112 | 5'UTR | 1 | T->G:1 | intergenic_variant | - | - | MODIFIER |
| 2019-nCoV_119 | 5'UTR | 1 | C->G:1 | intergenic_variant | - | - | MODIFIER |
| 2019-nCoV_120 | 5'UTR | 1 | T->C:1 | intergenic_variant | - | - | MODIFIER |
| 2019-nCoV_124 | 5'UTR | 1 | G->A:1 | intergenic_variant | - | - | MODIFIER |
| 2019-nCoV_241 | 5'UTR | 1 | C->T:1 | upstream_gene_variant | QHD43415.1 | gene-orf1ab | MODIFIER;DISTANCE=25 |
| 2019-nCoV_358 | gene-orf1ab | 1 | TGGAGACTCCGTGGAGGAGGTCTTA->T:1 | inframe_deletion | QHD43415.1:p.32-39GDSVEEVL>- | gene-orf1ab:c.94-117GGAGACTCCGTGGAGGAGGTCTTA>- | MODERATE |
| 2019-nCoV_490 | gene-orf1ab | 1 | T->W:1 | coding_sequence_variant | QHD43415.1:p.75- | gene-orf1ab:c.225gaT>gaW | MODIFIER |
| 2019-nCoV_583 | gene-orf1ab | 1 | C->T:1 | synonymous_variant | QHD43415.1:p.106V | gene-orf1ab:c.318gtC>gtT | LOW |
| 2019-nCoV_709 | gene-orf1ab | 1 | G->A:1 | synonymous_variant | QHD43415.1:p.148E | gene-orf1ab:c.444gaG>gaA | LOW |
| 2019-nCoV_1548 | gene-orf1ab | 1 | G->A:1 | missense_variant | QHD43415.1:p.428S>N | gene-orf1ab:c.1283aGc>aAc | MODERATE |
| 2019-nCoV_1912 | gene-orf1ab | 1 | C->T:1 | synonymous_variant | QHD43415.1:p.549S | gene-orf1ab:c.1647tcC>tcT | LOW |
| 2019-nCoV_3037 | gene-orf1ab | 1 | C->T:1 | synonymous_variant | QHD43415.1:p.924F | gene-orf1ab:c.2772ttC>ttT | LOW |
| 2019-nCoV_3177 | gene-orf1ab | 1 | C->Y:1 | coding_sequence_variant | QHD43415.1:p.971- | gene-orf1ab:c.2912cCt>cYt | MODIFIER |
| 2019-nCoV_3778 | gene-orf1ab | 1 | A->G:1 | synonymous_variant | QHD43415.1:p.1171T | gene-orf1ab:c.3513acA>acG | LOW |
| 2019-nCoV_4402 | gene-orf1ab | 1 | T->C:1 | synonymous_variant | QHD43415.1:p.1379L | gene-orf1ab:c.4137ctT>ctC | LOW |
| 2019-nCoV_5062 | gene-orf1ab | 1 | G->T:1 | missense_variant | QHD43415.1:p.1599L>F | gene-orf1ab:c.4797ttG>ttT | MODERATE |
| 2019-nCoV_6846 | gene-orf1ab | 1 | T->C:1 | missense_variant | QHD43415.1:p.2194M>T | gene-orf1ab:c.6581aTg>aCg | MODERATE |
| 2019-nCoV_6968 | gene-orf1ab | 1 | C->A:1 | missense_variant | QHD43415.1:p.2235L>I | gene-orf1ab:c.6703Cta>Ata | MODERATE |
| 2019-nCoV_6996 | gene-orf1ab | 1 | T->C:1 | missense_variant | QHD43415.1:p.2244I>T | gene-orf1ab:c.6731aTc>aCc | MODERATE |
| 2019-nCoV_7016 | gene-orf1ab | 1 | G->A:1 | missense_variant | QHD43415.1:p.2251G>S | gene-orf1ab:c.6751Ggt>Agt | MODERATE |
| 2019-nCoV_7866 | gene-orf1ab | 1 | G->T:1 | missense_variant | QHD43415.1:p.2534G>V | gene-orf1ab:c.7601gGt>gTt | MODERATE |
| 2019-nCoV_8001 | gene-orf1ab | 1 | A->C:1 | missense_variant | QHD43415.1:p.2579D>A | gene-orf1ab:c.7736gAt>gCt | MODERATE |
| 2019-nCoV_8388 | gene-orf1ab | 1 | A->G:1 | missense_variant | QHD43415.1:p.2708N>S | gene-orf1ab:c.8123aAc>aGc | MODERATE |
| 2019-nCoV_8782 | gene-orf1ab | 16 | C->T:15;C->Y:1 | synonymous_variant;coding_sequence_variant | QHD43415.1:p.2839S;QHD43415.1:p.2839- | gene-orf1ab:c.8517agC>agT;gene-orf1ab:c.8517agC>agY | LOW;MODIFIER |
| 2019-nCoV_8987 | gene-orf1ab | 1 | T->A:1 | missense_variant | QHD43415.1:p.2908F>I | gene-orf1ab:c.8722Ttt>Att | MODERATE |
| 2019-nCoV_9534 | gene-orf1ab | 1 | C->T:1 | missense_variant | QHD43415.1:p.3090T>I | gene-orf1ab:c.9269aCt>aTt | MODERATE |
| 2019-nCoV_9561 | gene-orf1ab | 1 | C->T:1 | missense_variant | QHD43415.1:p.3099S>L | gene-orf1ab:c.9296tCa>tTa | MODERATE |
| 2019-nCoV_11083 | gene-orf1ab | 1 | G->T:1 | missense_variant | QHD43415.1:p.3606L>F | gene-orf1ab:c.10818ttG>ttT | MODERATE |
| 2019-nCoV_11707 | gene-orf1ab | 1 | A->G:1 | synonymous_variant | QHD43415.1:p.3814L | gene-orf1ab:c.11442ttA>ttG | LOW |
| 2019-nCoV_11764 | gene-orf1ab | 1 | T->A:1 | missense_variant | QHD43415.1:p.3833N>K | gene-orf1ab:c.11499aaT>aaA | MODERATE |
| 2019-nCoV_15324 | gene-orf1ab | 1 | C->T:1 | synonymous_variant | QHD43415.1:p.5020N | gene-orf1ab:c.15060aaC>aaT | LOW |
| 2019-nCoV_15607 | gene-orf1ab | 1 | T->C:1 | synonymous_variant | QHD43415.1:p.5115L | gene-orf1ab:c.15343Tta>Cta | LOW |
| 2019-nCoV_16188 | gene-orf1ab | 1 | G->T:1 | missense_variant | QHD43415.1:p.5308W>C | gene-orf1ab:c.15924tgG>tgT | MODERATE |
| 2019-nCoV_17000 | gene-orf1ab | 1 | C->T:1 | missense_variant | QHD43415.1:p.5579T>I | gene-orf1ab:c.16736aCa>aTa | MODERATE |
| 2019-nCoV_17373 | gene-orf1ab | 2 | C->T:2 | synonymous_variant | QHD43415.1:p.5703A | gene-orf1ab:c.17109gcC>gcT | LOW |
| 2019-nCoV_18060 | gene-orf1ab | 3 | C->T:3 | synonymous_variant | QHD43415.1:p.5932L | gene-orf1ab:c.17796ctC>ctT | LOW |
| 2019-nCoV_18488 | gene-orf1ab | 2 | T->C:2 | missense_variant | QHD43415.1:p.6075I>T | gene-orf1ab:c.18224aTa>aCa | MODERATE |
| 2019-nCoV_18512 | gene-orf1ab | 1 | C->T:1 | missense_variant | QHD43415.1:p.6083P>L | gene-orf1ab:c.18248cCt>cTt | MODERATE |
| 2019-nCoV_19065 | gene-orf1ab | 1 | T->C:1 | synonymous_variant | QHD43415.1:p.6267P | gene-orf1ab:c.18801ccT>ccC | LOW |
| 2019-nCoV_19959 | gene-orf1ab | 1 | A->C:1 | missense_variant | QHD43415.1:p.6565E>D | gene-orf1ab:c.19695gaA>gaC | MODERATE |
| 2019-nCoV_20670 | gene-orf1ab | 2 | G->A:2 | synonymous_variant | QHD43415.1:p.6802A | gene-orf1ab:c.20406gcG>gcA | LOW |
| 2019-nCoV_20679 | gene-orf1ab | 2 | G->A:2 | synonymous_variant | QHD43415.1:p.6805P | gene-orf1ab:c.20415ccG>ccA | LOW |
| 2019-nCoV_21137 | gene-orf1ab | 1 | A->G:1 | missense_variant | QHD43415.1:p.6958K>R | gene-orf1ab:c.20873aAg>aGg | MODERATE |
| 2019-nCoV_21316 | gene-orf1ab | 1 | G->A:1 | missense_variant | QHD43415.1:p.7018D>N | gene-orf1ab:c.21052Gat>Aat | MODERATE |
| 2019-nCoV_21656 | gene-S | 1 | T->A:1 | missense_variant | QHD43416.1:p.32F>I | gene-S:c.94Ttc>Atc | MODERATE |
| 2019-nCoV_21707 | gene-S | 3 | C->T:3 | missense_variant | QHD43416.1:p.49H>Y | gene-S:c.145Cat>Tat | MODERATE |
| 2019-nCoV_22303 | gene-S | 1 | T->G:1 | missense_variant | QHD43416.1:p.247S>R | gene-S:c.741agT>agG | MODERATE |
| 2019-nCoV_22586 | gene-S | 1 | T->Y:1 | coding_sequence_variant | QHD43416.1:p.342- | gene-S:c.1024Ttt>Ytt | MODIFIER |
| 2019-nCoV_22622 | gene-S | 1 | A->G:1 | missense_variant | QHD43416.1:p.354N>D | gene-S:c.1060Aac>Gac | MODERATE |
| 2019-nCoV_22652 | gene-S | 1 | G->T:1 | missense_variant | QHD43416.1:p.364D>Y | gene-S:c.1090Gat>Tat | MODERATE |
| 2019-nCoV_22661 | gene-S | 2 | G->T:2 | missense_variant | QHD43416.1:p.367V>F | gene-S:c.1099Gtc>Ttc | MODERATE |
| 2019-nCoV_23403 | gene-S | 1 | A->G:1 | missense_variant | QHD43416.1:p.614D>G | gene-S:c.1841gAt>gGt | MODERATE |
| 2019-nCoV_23569 | gene-S | 2 | T->C:2 | synonymous_variant | QHD43416.1:p.669G | gene-S:c.2007ggT>ggC | LOW |
| 2019-nCoV_23605 | gene-S | 2 | T->G:2 | synonymous_variant | QHD43416.1:p.681P | gene-S:c.2043ccT>ccG | LOW |
| 2019-nCoV_24034 | gene-S | 2 | C->T:1;C->Y:1 | synonymous_variant;coding_sequence_variant | QHD43416.1:p.824N;QHD43416.1:p.824- | gene-S:c.2472aaC>aaT;gene-S:c.2472aaC>aaY | LOW;MODIFIER |
| 2019-nCoV_24325 | gene-S | 2 | A->G:2 | synonymous_variant | QHD43416.1:p.921K | gene-S:c.2763aaA>aaG | LOW |
| 2019-nCoV_25060 | gene-S | 1 | A->G:1 | synonymous_variant | QHD43416.1:p.1166L | gene-S:c.3498ttA>ttG | LOW |
| 2019-nCoV_25645 | gene-ORF3a | 1 | T->C:1 | synonymous_variant | QHD43417.1:p.85L | gene-ORF3a:c.253Ttg>Ctg | LOW |
| 2019-nCoV_25964 | gene-ORF3a | 1 | A->G:1 | missense_variant | QHD43417.1:p.191E>G | gene-ORF3a:c.572gAa>gGa | MODERATE |
| 2019-nCoV_26144 | gene-ORF3a | 5 | G->T:5 | missense_variant | QHD43417.1:p.251G>V | gene-ORF3a:c.752gGt>gTt | MODERATE |
| 2019-nCoV_26729 | gene-M | 2 | T->C:1;T->Y:1 | synonymous_variant;coding_sequence_variant | QHD43419.1:p.69A;QHD43419.1:p.69- | gene-M:c.207gcT>gcC;gene-M:c.207gcT>gcY | LOW;MODIFIER |
| 2019-nCoV_27493 | gene-ORF7a | 2 | C->T:2 | missense_variant | QHD43421.1:p.34P>S | gene-ORF7a:c.100Cct>Tct | MODERATE |
| 2019-nCoV_27577 | gene-ORF7a | 1 | C->T:1 | stop_gained | QHD43421.1:p.62Q>* | gene-ORF7a:c.184Caa>Taa | HIGH |
| 2019-nCoV_28077 | gene-ORF8 | 2 | G->S:1;G->C:1 | coding_sequence_variant;missense_variant | QHD43422.1:p.62-;QHD43422.1:p.62V>L | gene-ORF8:c.184Gtg>Stg;gene-ORF8:c.184Gtg>Ctg | MODIFIER;MODERATE |
| 2019-nCoV_28144 | gene-ORF8 | 16 | T->C:15;T->Y:1 | missense_variant;coding_sequence_variant | QHD43422.1:p.84L>S;QHD43422.1:p.84- | gene-ORF8:c.251tTa>tCa;gene-ORF8:c.251tTa>tYa | MODERATE;MODIFIER |
| 2019-nCoV_28253 | gene-ORF8 | 2 | C->T:2 | synonymous_variant | QHD43422.1:p.120F | gene-ORF8:c.360ttC>ttT | LOW |
| 2019-nCoV_28291 | gene-N | 1 | C->T:1 | synonymous_variant | QHD43423.2:p.6P | gene-N:c.18ccC>ccT | LOW |
| 2019-nCoV_28716 | gene-N | 1 | C->T:1 | missense_variant | QHD43423.2:p.148T>I | gene-N:c.443aCc>aTc | MODERATE |
| 2019-nCoV_28792 | gene-N | 1 | A->T:1 | synonymous_variant | QHD43423.2:p.173A | gene-N:c.519gcA>gcT | LOW |
| 2019-nCoV_28854 | gene-N | 3 | C->T:2;C->Y:1 | missense_variant;coding_sequence_variant | QHD43423.2:p.194S>L;QHD43423.2:p.194- | gene-N:c.581tCa>tTa;gene-N:c.581tCa>tYa | MODERATE;MODIFIER |
| 2019-nCoV_29095 | gene-N | 7 | C->T:7 | synonymous_variant | QHD43423.2:p.274F | gene-N:c.822ttC>ttT | LOW |
| 2019-nCoV_29303 | gene-N | 1 | C->T:1 | missense_variant | QHD43423.2:p.344P>S | gene-N:c.1030Cca>Tca | MODERATE |
| 2019-nCoV_29596 | gene-ORF10 | 1 | A->G:1 | missense_variant | QHI42199.1:p.13I>M | gene-ORF10:c.39atA>atG | MODERATE |
| 2019-nCoV_29749 | 3'UTR | 1 | ACGATCGAGTG->A:1 | downstream_gene_variant | QHI42199.1 | gene-ORF10 | MODIFIER;DISTANCE=76 |
| 2019-nCoV_29854 | 3'UTR | 1 | C->T:1 | intergenic_variant | - | - | MODIFIER |
| 2019-nCoV_29856 | 3'UTR | 1 | T->A:1 | intergenic_variant | - | - | MODIFIER |
| 2019-nCoV_29868 | 3'UTR | 1 | GA->G:1 | intergenic_variant | - | - | MODIFIER |
| 2019-nCoV_29877 | 3'UTR | 1 | A->T:1 | intergenic_variant | - | - | MODIFIER |
参考文献
Novel Coronavirus (2019-nCoV)
https://www.who.int/emergencies/diseases/novel-coronavirus-2019,
Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission
Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin
Homologous recombination within the spike glycoprotein of the newly identified coronavirus may boost cross-species transmission from snake to human
Genomic and protein structure modelling analysis depicts the origin and infectivity of 2019-nCoV, a new coronavirus which caused a pneumonia outbreak in Wuhan, China
The 2019-new Coronavirus epidemic: evidence for virus evolution
Bioinformatics analysis of the Wuhan 2019 human coronavirus genome
武汉2019冠状病毒基因组的生物信息学分析
Data sharing and outbreaks: best practice exemplified
Koopmans M, van Doremalen N, van Riel D, de Wit E. A novel coronavirus emerging in China - key questions for impact assessment
Database resources of the National Center for Biotechnology Information
GISAID: Global initiative on sharing all influenza data-from vision to reality
The China National GeneBank—owned by all, completed by all and shared by all
国家基因库: 共有、共为、共享
World data centre for microorganisms: an information infrastructure to explore and utilize preserved microbial strains worldwide
Database resources of the National Genomics Data Center in 2020
GSA: genome sequence archive
GSA: genome sequence archive
GSA: 组学原始数据归档库
The BIG Data Center’s database resources
生命与健康大数据中心资源
Prospects for national biological big data centers
国家级生物大数据中心展望
MUSCLE: multiple sequence alignment with high accuracy and high throughput
Hochreiter S. msa: an R package for multiple sequence alignment
Using the generic genome browser(GBrowse)
.The ensembl variant effect predictor








 南京肽业生物科技有限公司
南京肽业生物科技有限公司
 地址:
地址:
 Email: info@njpeptide.com
Email: info@njpeptide.com
 总机:025-58361106-801
总机:025-58361106-801
